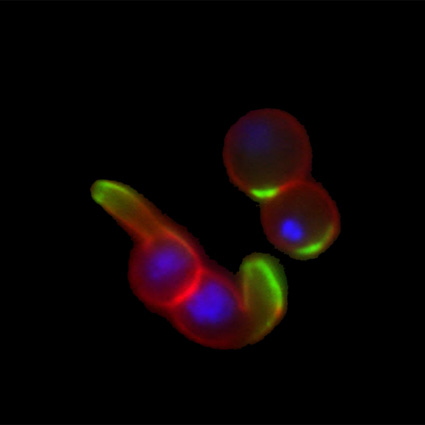
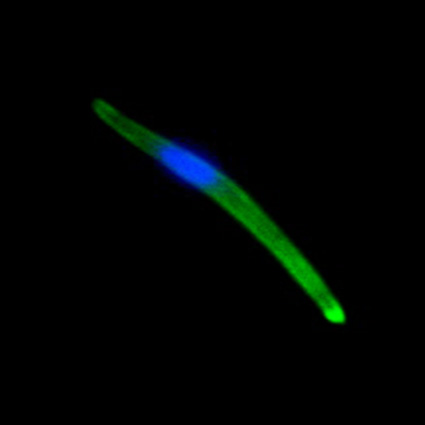
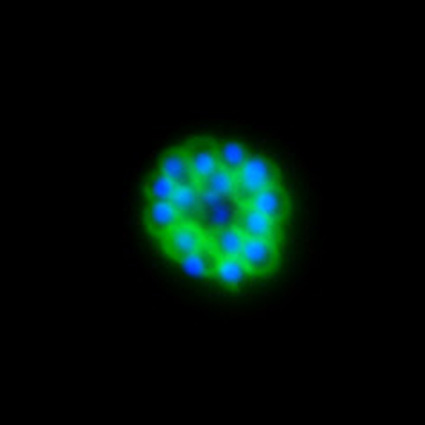
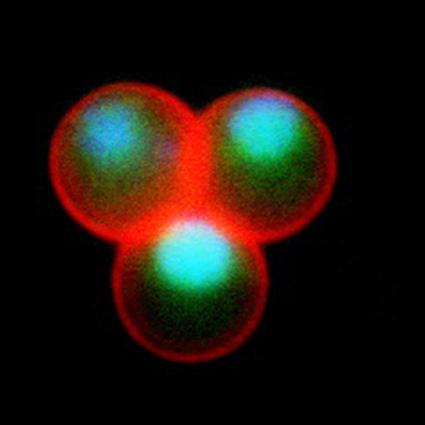
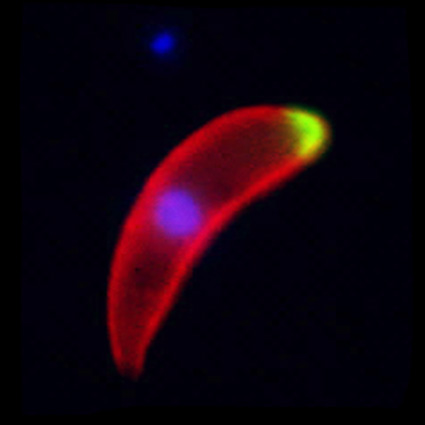
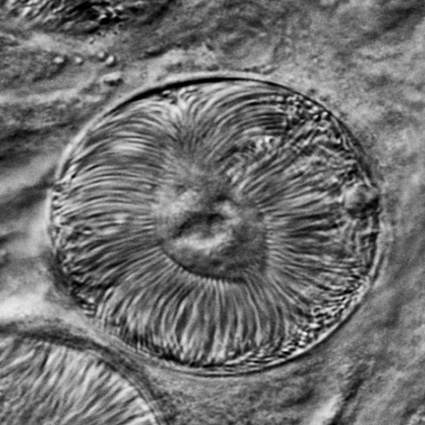
Malaria is an infectious disease that threatens 40% of total world’s population, the majority living in developing countries. There were an estimated 241 million malaria cases and 627 000 malaria deaths worldwide in 2020. The disease is caused by an apicomplexan protozoan parasite of the genus Plasmodium that is transmitted by the bite of an infected female Anopheles mosquito. The parasite develops and multiplies within the liver of the human host and then infects and multiplies within red blood cells. This asexual blood stage is responsible for the pathology of the disease. The control of the disease is hampered by the lack of an effective vaccine and the ever increasing resistance to antiparasite drugs and of mosquitoes to insecticides. Control strategies are further limited by the complex life cycle of the malaria parasite that has developmental stages within the liver and blood of the human host and in the gut and salivary glands of the mosquito. To develop rational and effective intervention strategies, understanding the biology of parasite development and the signalling processes that control it are crucial.
Our research is mainly aimed to understand the molecular pathways that are involved in malaria parasite development and differentiation, cell division, and parasite interactions with both mammalian and vector host (https://www.nottingham.ac.uk/life-sciences/people/rita.tewari). We use molecular, genetic, cell biology, biochemistry, proteomics and reverse genetics approaches to study the function of regulatory protein families like kinases, phosphatases, cell division proteins and armadillo repeat proteins in malaria parasite biology. The aim is to identify the best drug and vaccine targets and along the way understand basic parasite cell and developmental biology. For all these studies we use rodent malaria model Plasmodium berghei that is easily amenable to reverse genetic studies and whole life cycle of parasite can be studied both in mammalian host and vector mosquito.